Group leader: Prof. dr. Marcel Kool
The research group of Prof. dr. Marcel Kool studies the genomics and epigenomics of pediatric brain tumors and how to translate findings from these studies into novel therapies. More effective and less toxic therapies are highly needed as there are many types of childhood brain tumors for which the survival is still very poor or where survivors suffer from serious long-term side effects caused by their intensive therapies. In order to develop such new therapies, we need many preclinical models, molecularly well-characterized, and representing the broad spectrum of different pediatric brain tumor entities and the inter-tumor heterogeneity within each of these entities. We will use these models to get a better understanding of tumor origin and biology and to find new ways how to target them.
The types of pediatric brain tumors that the group is mostly focusing on are ependymomas (EPN) and embryonal brain tumors. The latter include medulloblastomas (MB), atypical / teratoid rhabdoid tumors (ATRT), embryonal tumors with multilayered rosettes (ETMR), and other tumor types previously called primitive neuroectodermal tumors of the central nervous system (CNS-PNETs). PI Marcel Kool is a world leading expert in the classification and molecular characterization of these types of tumors and has published several landmark papers for each of these different entities.
We work in close collaboration with the other research team that Prof. dr. Marcel Kool leads at the Hopp Children’s Cancer Center (KiTZ) in Heidelberg, Germany. Both team are part of these same research program and focus on different aspects of ependymomas and embryonal brain tumors. The group in Heidelberg works on the genomic and epigenomic analyses of these types of tumors and how to translate (epi)genomic findings to the clinic using patient-derived xenograft (PDX) mouse models. They are also strongly involved in the ITCC-P4 consortium that aims to establish and characterize >400 PDX models of solid pediatric cancers and use these for high throughput in vivo preclinical studies. The group in Utrecht is modeling these tumors using organoid technology to better understand the biology of these tumors and to perform preclinical experiments. Combining the complementary expertise, data and other knowledge from both groups and both centers helps to accelerate our research.
Understanding what is driving tumor development and growth
An improved molecular classification and characterization of pediatric brain tumors has uncovered many distinct types of brain tumors, each harboring their own specific pattern of mutations and genomic aberrations. Although likely representing tumor-driving events, we do not know why or how these genetic changes cause tumor formation. However, this knowledge is essential for understand tumor biology.
Embryonal brain tumors and ependymomas arise from progenitor cells during brain development, although we are still determining the precise cell-of-origin and timing for each of these entities. Consequently, the tumors resemble more the immature progenitor cells than mature neurons or glia, and we need to understand what keeps these cells growing and prevents them from differentiating. Hence, we need to investigate the tumor specific genetic changes in the context of these immature progenitor cells. Brain organoids, or “mini-brains in a Petri dish” provide an ideal in vitro model for this research question. By using different protocols, the brain organoids can mimic in 3D the various steps of normal brain development of different regions of the central nervous system, including the cerebral cortex and the cerebellum. By using reprogrammed healthy cells from a patient and targeted gene manipulation, we are able to imitate what happens in each individual patient and follow the formation of a tumor. We can then analyze these genetically engineered brain tumor organoids at different stages of their development using state-of-the-art technologies. For instance, we can determine how tumor cells change over time and differ from normal progenitors by single cell sequencing and lineage tracing. We can also compare how different mutations lead to different tumors and identify the precise cell-of-origin. Together, these experiments will provide a unique window in the early events of tumor development.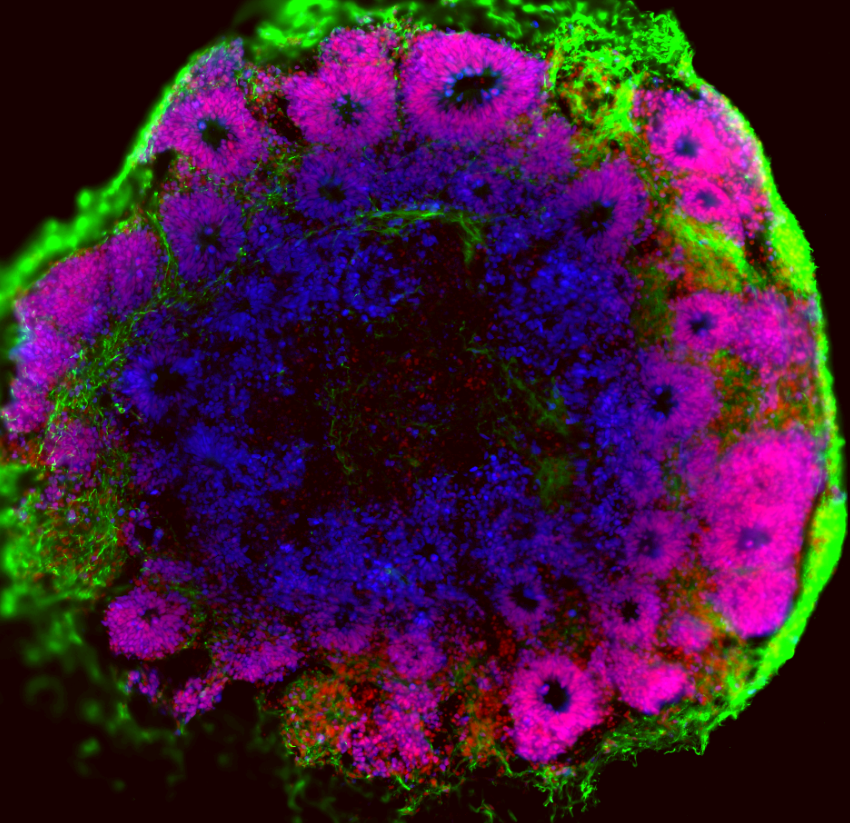
Tumor organoids as personalized preclinical models
The recent discovery of many different subtypes and tumor entities also revealed a potential problem for finding more efficient treatments: not all tumors are alike! Therefore, we need many new and well-characterized preclinical models representing all the different molecular subtypes to test new therapeutic strategies. Together with other groups at the Princess Máxima Center, we are generating brain tumor organoids (BTOs). These are 3D cultures of tumors from our patients. This BTO approach has advantages over other approaches. Compared to normal 2D cell culture techniques, these BTOs better represent the original tumor and its heterogeneity. It also allows a more high-throughput approach than using tumor xenografts in mice. We aim to generate a living biobank of patient derived BTOs. We will use these organoids in drug screens to investigate whether the tumor might be sensitive for FDA approved drugs and new drug candidates.
Tumors always exist within the context healthy brain cells. To study this and to better understand the relationship between tumor cells and the surrounding normal brain tissue, we will combine tumor organoids with normal brain organoids. These brain organoid – tumor co-cultures enable us to screen for drugs that specifically target the tumor cells and not the normal brain cells. We will also us this model to study how tumor cells migrate and metastasize.
Grants
- Modelling Embryonal Brain Tumors with BCOR Alterations for Understanding Biology and Therapy - KiKa project grant (2024 - 2028)
- Improving treatments for patients with rare embryonal and sarcomatous CNS tumours – Quest for Cures: Collaborative Discovery Teams program grant – The Brain Tumour Charity UK (2024 - 2029) – with Katja Hoff von Hoff (Aarhus), Barry Pizer (Liverpool), Darren Hargrave (London), Jan Koster (Amsterdam) and Johannes Gojo (Vienna)
- MIMIC 2.0 – Myeloid cell targeting to enhance T-cell immunotherapy in pediatric high-grade brain cancers - KiKa project grant (2024 - 2028) - with Jasper van der Lugt, Nierkens Group and Stunnenberg Group
- Ependymoma models and drug screening - Ependymoma Research Foundation (2024-2025)
- Generation, characterization and testing of new preclinical models for rare brain tumor types - Oncode Accelerator Brain Program (2024-2026)
- Exploring Unknown Relapse Origins in Paediatric Ependymoma (EUROPE) – Fight Kids Cancer project grant (2024 - 2028) – with Kristian Pajtler (KiTZ), Abigail Suwala (Heidelberg) and Ülrich Schüller (Hamburg) and Johannes Gojo (Vienna)
- The ITCC Brain Translational Accelerator Platform – Fight Kids Cancer project grant (2024 - 2028) – with David Jones / Kristian Pajtler (KiTZ), Steve Clifford/Gareth Veal (Newcastle), Chris Jones (London) and Johannes Gojo (Vienna)
- Cerebellar Organoid Modelling of Medulloblastoma to Accelerate New Discoveries (COMMAND) – Quest for Cures: Collaborative Discovery Teams program grant – The Brain Tumour Charity UK (2023 - 2028) – with Esther Becker (NCDN, Oxford), Lena Kütscher (KiTZ, Heidelberg) and Paul Northcott (St Jude, Memphis)
- Genetically modified cerebellar organoids as models for sonic hedgehog medulloblastoma - KiKa project grant (2022 - 2025)
- High-throughput drug screens on PDX-derived organoids for posterior fossa A ependymoma – Stichting STOPHersentumoren.nl (2022 - 2023) – News article
- ITCC-P4 MIRROR project with the Molenaar Group and other groups in the Máxima
- Marie Sklodowska-Curie COFUND – Máxima Butterfly Program with 25 other research groups in the Máxima –News article
- Brain tumor organoids as models for pediatric brain tumors - KiKa fast track grant (2019 - 2023)
Twinning program KiTZ
- Personalized drug sensitivity profiling– with Molenaar Group and other groups in KiTZ and the Máxima
- KITZ-Máxima PDX pipeline – with various groups in KiTZ and the Máxima
D.T. Jones*, N. Jager*, M. Kool, T. Zichner, B. Hutter, M. Sultan, Y.J. Cho, T.J. Pugh, V. Hovestadt, A.M. Stutz, T. Rausch, H.J. Warnatz, M. Ryzhova, S. Bender, D. Sturm, S. Pleier, H. Cin, E. Pfaff, L. Sieber, A. Wittmann, M. Remke, H. Witt, S. Hutter, T. Tzaridis, J. Weischenfeldt, B. Raeder, M. Avci, V. Amstislavskiy, M. Zapatka, U.D. Weber, Q. Wang, B. Lasitschka, C.C. Bartholomae, M. Schmidt, C. von Kalle, V. Ast, C. Lawerenz, J. Eils, R. Kabbe, V. Benes, P. van Sluis, J. Koster, R. Volckmann, D. Shih, M.J. Betts, R.B. Russell, S. Coco, G.P. Tonini, U. Schuller, V. Hans, N. Graf, Y.J. Kim, C. Monoranu, W. Roggendorf, A. Unterberg, C. Herold-Mende, T. Milde, A.E. Kulozik, A. von Deimling, O. Witt, E. Maass, J. Rossler, M. Ebinger, M.U. Schuhmann, M.C. Fruhwald, M. Hasselblatt, N. Jabado, S. Rutkowski, A.O. von Bueren, D. Williamson, S.C. Clifford, M.G. McCabe, V.P. Collins, S. Wolf, S. Wiemann, H. Lehrach, B. Brors, W. Scheurlen, J. Felsberg, G. Reifenberger, P.A. Northcott, M.D. Taylor, M. Meyerson, S.L. Pomeroy, M.L. Yaspo, J.O. Korbel, A. Korshunov, R. Eils*#, S.M. Pfister*#, P. Lichter*, Dissecting the genomic complexity underlying medulloblastoma, Nature, 488 (2012) 100-105.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3662966/
Kool#, D.T. Jones, N. Jager, P.A. Northcott, T.J. Pugh, V. Hovestadt, R.M. Piro, L.A. Esparza, S.L. Markant, M. Remke, T. Milde, F. Bourdeaut, M. Ryzhova, D. Sturm, E. Pfaff, S. Stark, S. Hutter, H. Seker-Cin, P. Johann, S. Bender, C. Schmidt, T. Rausch, D. Shih, J. Reimand, L. Sieber, A. Wittmann, L. Linke, H. Witt, U.D. Weber, M. Zapatka, R. Konig, R. Beroukhim, G. Bergthold, P. van Sluis, R. Volckmann, J. Koster, R. Versteeg, S. Schmidt, S. Wolf, C. Lawerenz, C.C. Bartholomae, C. von Kalle, A. Unterberg, C. Herold-Mende, S. Hofer, A.E. Kulozik, A. von Deimling, W. Scheurlen, J. Felsberg, G. Reifenberger, M. Hasselblatt, J.R. Crawford, G.A. Grant, N. Jabado, A. Perry, C. Cowdrey, S. Croul, G. Zadeh, J.O. Korbel, F. Doz, O. Delattre, G.D. Bader, M.G. McCabe, V.P. Collins, M.W. Kieran, Y.J. Cho, S.L. Pomeroy, O. Witt, B. Brors, M.D. Taylor, U. Schuller, A. Korshunov, R. Eils, R.J. Wechsler-Reya*, P. Lichter*, S.M. Pfister*, I.P.T. Project, Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition, Cancer Cell, 25 (2014) 393-405.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4493053/
K.W. Pajtler*, H. Witt*, M. Sill*, D.T. Jones, V. Hovestadt, F. Kratochwil, K. Wani, R. Tatevossian, C. Punchihewa, P. Johann, J. Reimand, H.J. Warnatz, M. Ryzhova, S. Mack, V. Ramaswamy, D. Capper, L. Schweizer, L. Sieber, A. Wittmann, Z. Huang, P. van Sluis, R. Volckmann, J. Koster, R. Versteeg, D. Fults, H. Toledano, S. Avigad, L.M. Hoffman, A.M. Donson, N. Foreman, E. Hewer, K. Zitterbart, M. Gilbert, T.S. Armstrong, N. Gupta, J.C. Allen, M.A. Karajannis, D. Zagzag, M. Hasselblatt, A.E. Kulozik, O. Witt, V.P. Collins, K. von Hoff, S. Rutkowski, T. Pietsch, G. Bader, M.L. Yaspo, A. von Deimling, P. Lichter, M.D. Taylor, R. Gilbertson, D.W. Ellison, K. Aldape, A. Korshunov*, M. Kool*#, S.M. Pfister*#, Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups, Cancer Cell, 27 (2015) 728-743.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4712639/
P.D. Johann*, S. Erkek*, M. Zapatka, K. Kerl, I. Buchhalter, V. Hovestadt, D.T. Jones, D. Sturm, C. Hermann, M. Segura Wang, A. Korshunov, M. Rhyzova, S. Grobner, S. Brabetz, L. Chavez, S. Bens, S. Groschel, F. Kratochwil, A. Wittmann, L. Sieber, C. Georg, S. Wolf, K. Beck, F. Oyen, D. Capper, P. van Sluis, R. Volckmann, J. Koster, R. Versteeg, A. von Deimling, T. Milde, O. Witt, A.E. Kulozik, M. Ebinger, T. Shalaby, M. Grotzer, D. Sumerauer, J. Zamecnik, J. Mora, N. Jabado, M.D. Taylor, A. Huang, E. Aronica, A. Bertoni, B. Radlwimmer, T. Pietsch, U. Schuller, R. Schneppenheim, P.A. Northcott, J.O. Korbel, R. Siebert, M.C. Fruhwald, P. Lichter, R. Eils, A. Gajjar, M. Hasselblatt, S.M. Pfister*, M. Kool*#, Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes, Cancer Cell, 29 (2016) 379-393.
https://www.cell.com/cancer-cell/fulltext/S1535-6108(16)30035-6
Sturm*, B.A. Orr*, U.H. Toprak*, V. Hovestadt*, D.T. Jones, D. Capper, M. Sill, I. Buchhalter, P.A. Northcott, I. Leis, M. Ryzhova, C. Koelsche, E. Pfaff, S.J. Allen, G. Balasubramanian, B.C. Worst, K.W. Pajtler, S. Brabetz, P.D. Johann, F. Sahm, J. Reimand, A. Mackay, D.M. Carvalho, M. Remke, J.J. Phillips, A. Perry, C. Cowdrey, R. Drissi, M. Fouladi, F. Giangaspero, M. Lastowska, W. Grajkowska, W. Scheurlen, T. Pietsch, C. Hagel, J. Gojo, D. Lotsch, W. Berger, I. Slavc, C. Haberler, A. Jouvet, S. Holm, S. Hofer, M. Prinz, C. Keohane, I. Fried, C. Mawrin, D. Scheie, B.C. Mobley, M.J. Schniederjan, M. Santi, A.M. Buccoliero, S. Dahiya, C.M. Kramm, A.O. von Bueren, K. von Hoff, S. Rutkowski, C. Herold-Mende, M.C. Fruhwald, T. Milde, M. Hasselblatt, P. Wesseling, J. Rossler, U. Schuller, M. Ebinger, J. Schittenhelm, S. Frank, R. Grobholz, I. Vajtai, V. Hans, R. Schneppenheim, K. Zitterbart, V.P. Collins, E. Aronica, P. Varlet, S. Puget, C. Dufour, J. Grill, D. Figarella-Branger, M. Wolter, M.U. Schuhmann, T. Shalaby, M. Grotzer, T. van Meter, C.M. Monoranu, J. Felsberg, G. Reifenberger, M. Snuderl, L.A. Forrester, J. Koster, R. Versteeg, R. Volckmann, P. van Sluis, S. Wolf, T. Mikkelsen, A. Gajjar, K. Aldape, A.S. Moore, M.D. Taylor, C. Jones, N. Jabado, M.A. Karajannis, R. Eils, M. Schlesner, P. Lichter, A. von Deimling, S.M. Pfister, D.W. Ellison*, A. Korshunov*, M. Kool*#, New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs, Cell, 164 (2016) 1060-1072.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5139621/
S.C. Mack*, K.W. Pajtler*, L. Chavez*, K. Okonechnikov, K.C. Bertrand, X. Wang, S. Erkek, A. Federation, A. Song, C. Lee, X. Wang, L. McDonald, J.J. Morrow, A. Saiakhova, P. Sin-Chan, Q. Wu, K.A. Michaelraj, T.E. Miller, C.G. Hubert, M. Ryzhova, L. Garzia, L. Donovan, S. Dombrowski, D.C. Factor, B. Luu, C.L.L. Valentim, R.C. Gimple, A. Morton, L. Kim, B.C. Prager, J.J.Y. Lee, X. Wu, J. Zuccaro, Y. Thompson, B.L. Holgado, J. Reimand, S.Q. Ke, A. Tropper, S. Lai, S. Vijayarajah, S. Doan, V. Mahadev, A.F. Minan, S.N. Grobner, M. Lienhard, M. Zapatka, Z. Huang, K.D. Aldape, A.M. Carcaboso, P.J. Houghton, S.T. Keir, T. Milde, H. Witt, Y. Li, C.J. Li, X.W. Bian, D.T.W. Jones, I. Scott, S.K. Singh, A. Huang, P.B. Dirks, E. Bouffet, J.E. Bradner, V. Ramaswamy, N. Jabado, J.T. Rutka, P.A. Northcott, M. Lupien, P. Lichter, A. Korshunov, P.C. Scacheri, S.M. Pfister, M. Kool*#, M.D. Taylor*#, J.N. Rich*#, Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling, Nature, 553 (2018) 101-105.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5993422/
Capper*, D.T.W. Jones*, M. Sill*, V. Hovestadt*, D. Schrimpf, D. Sturm, C. Koelsche, F. Sahm, L. Chavez, D.E. Reuss, A. Kratz, A.K. Wefers, K. Huang, K.W. Pajtler, L. Schweizer, D. Stichel, A. Olar, N.W. Engel, K. Lindenberg, P.N. Harter, A.K. Braczynski, K.H. Plate, H. Dohmen, B.K. Garvalov, R. Coras, A. Holsken, E. Hewer, M. Bewerunge-Hudler, M. Schick, R. Fischer, R. Beschorner, J. Schittenhelm, O. Staszewski, K. Wani, P. Varlet, M. Pages, P. Temming, D. Lohmann, F. Selt, H. Witt, T. Milde, O. Witt, E. Aronica, F. Giangaspero, E. Rushing, W. Scheurlen, C. Geisenberger, F.J. Rodriguez, A. Becker, M. Preusser, C. Haberler, R. Bjerkvig, J. Cryan, M. Farrell, M. Deckert, J. Hench, S. Frank, J. Serrano, K. Kannan, A. Tsirigos, W. Bruck, S. Hofer, S. Brehmer, M. Seiz-Rosenhagen, D. Hanggi, V. Hans, S. Rozsnoki, J.R. Hansford, P. Kohlhof, B.W. Kristensen, M. Lechner, B. Lopes, C. Mawrin, R. Ketter, A. Kulozik, Z. Khatib, F. Heppner, A. Koch, A. Jouvet, C. Keohane, H. Muhleisen, W. Mueller, U. Pohl, M. Prinz, A. Benner, M. Zapatka, N.G. Gottardo, P.H. Driever, C.M. Kramm, H.L. Muller, S. Rutkowski, K. von Hoff, M.C. Fruhwald, A. Gnekow, G. Fleischhack, S. Tippelt, G. Calaminus, C.M. Monoranu, A. Perry, C. Jones, T.S. Jacques, B. Radlwimmer, M. Gessi, T. Pietsch, J. Schramm, G. Schackert, M. Westphal, G. Reifenberger, P. Wesseling, M. Weller, V.P. Collins, I. Blumcke, M. Bendszus, J. Debus, A. Huang, N. Jabado, P.A. Northcott, W. Paulus, A. Gajjar, G.W. Robinson, M.D. Taylor, Z. Jaunmuktane, M. Ryzhova, M. Platten, A. Unterberg, W. Wick, M.A. Karajannis, M. Mittelbronn, T. Acker, C. Hartmann, K. Aldape, U. Schuller, R. Buslei, P. Lichter, M. Kool, C. Herold-Mende, D.W. Ellison, M. Hasselblatt, M. Snuderl, S. Brandner, A. Korshunov, A. von Deimling*#, S.M. Pfister*#, DNA methylation-based classification of central nervous system tumours, Nature, 555 (2018) 469-474.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6093218/
S.N. Grobner*, B.C. Worst*, J. Weischenfeldt, I. Buchhalter, K. Kleinheinz, V.A. Rudneva, P.D. Johann, G.P. Balasubramanian, M. Segura-Wang, S. Brabetz, S. Bender, B. Hutter, D. Sturm, E. Pfaff, D. Hubschmann, G. Zipprich, M. Heinold, J. Eils, C. Lawerenz, S. Erkek, S. Lambo, S. Waszak, C. Blattmann, A. Borkhardt, M. Kuhlen, A. Eggert, S. Fulda, M. Gessler, J. Wegert, R. Kappler, D. Baumhoer, S. Burdach, R. Kirschner-Schwabe, U. Kontny, A.E. Kulozik, D. Lohmann, S. Hettmer, C. Eckert, S. Bielack, M. Nathrath, C. Niemeyer, G.H. Richter, J. Schulte, R. Siebert, F. Westermann, J.J. Molenaar, G. Vassal, H. Witt, I.P.-S. Project, I.M.-S. Project, B. Burkhardt, C.P. Kratz, O. Witt, C.M. van Tilburg, C.M. Kramm, G. Fleischhack, U. Dirksen, S. Rutkowski, M. Fruhwald, K. von Hoff, S. Wolf, T. Klingebiel, E. Koscielniak, P. Landgraf, J. Koster, A.C. Resnick, J. Zhang, Y. Liu, X. Zhou, A.J. Waanders, D.A. Zwijnenburg, P. Raman, B. Brors, U.D. Weber, P.A. Northcott, K.W. Pajtler, M. Kool, R.M. Piro, J.O. Korbel, M. Schlesner, R. Eils, D.T.W. Jones, P. Lichter, L. Chavez*, M. Zapatka*, S.M. Pfister*#, The landscape of genomic alterations across childhood cancers, Nature, 555 (2018) 321-327.
https://www.nature.com/articles/nature25480
Lambo, S.N. Grobner, T. Rausch, S.M. Waszak, C. Schmidt, A. Gorthi, J.C. Romero, M. Mauermann, S. Brabetz, S. Krausert, I. Buchhalter, J. Koster, D.A. Zwijnenburg, M. Sill, J.M. Hubner, N. Mack, B. Schwalm, M. Ryzhova, V. Hovestadt, S. Papillon-Cavanagh, J.A. Chan, P. Landgraf, B. Ho, T. Milde, O. Witt, J. Ecker, F. Sahm, D. Sumerauer, D.W. Ellison, B.A. Orr, A. Darabi, C. Haberler, D. Figarella-Branger, P. Wesseling, J. Schittenhelm, M. Remke, M.D. Taylor, M.J. Gil-da-Costa, M. Lastowska, W. Grajkowska, M. Hasselblatt, P. Hauser, T. Pietsch, E. Uro-Coste, F. Bourdeaut, J. Masliah-Planchon, V. Rigau, S. Alexandrescu, S. Wolf, X.N. Li, U. Schuller, M. Snuderl, M.A. Karajannis, F. Giangaspero, N. Jabado, A. von Deimling, D.T.W. Jones, J.O. Korbel, K. von Hoff, P. Lichter, A. Huang, A.J.R. Bishop, S.M. Pfister*, A. Korshunov*, M. Kool*#, The molecular landscape of ETMR at diagnosis and relapse, Nature, 576 (2019) 274-280.
https://www.nature.com/articles/s41586-019-1815-x
* Equal contribution
# Corresponding author
More information
More about our group at KiTZ:
More publications of Marcel Kool:
https://scholar.google.nl/citations?user=8Xzec98AAAAJ&hl=en
Websites:
Job opportunities in our research department











