
Group leader: Dr. Jarno Drost
Cancer is still one of the leading causes of disease-related deaths in children. Survivors suffer from side effects of the, in most cases, intensive treatment regimens. Hence, there is an urgent need to develop improved, less toxic therapies. Our vision is that if we understand the processes that underpin the development of childhood cancer, we will facilitate improved cure rates and a better quality of life. However, therapeutic innovation is hampered by the lack of cell models representative of native tumor tissue. We are convinced that by developing cancer models that more closely recapitulate the patient's tumor, we will be able to more efficiently translate pre-clinical findings to the clinic and bridge the gap between bench and bedside: Better models, increased knowledge: more cures!
Organoids as representative models for childhood solid tumors
The organoid technology has revolutionized cancer research, as it allows for the ‘unlimited’ expansion of healthy and diseased tissue from individual patients in a dish, while retaining essential characteristics of native tissue. Organoids are therefore seen as avatars of the tissue they were derived from. The Drost group pioneered the use of organoid technology for pediatric cancer research. We succeeded in establishing culture protocols to grow organoids from a wide spectrum of pediatric malignancies such as Wilms tumors, renal cell carcinomas, as well as different rhabdoid tumor and soft tissue sarcoma subtypes. We use these models to study the very fundamental processes underlying tumorigenesis (see below), as well as for more translational research projects. For instance, we use the organoids as a drug screen platform to find tumor-specific drug vulnerabilities, but we also develop co-culture systems of organoids and different types of immune cells to explore the use of immunotherapy in pediatric cancer.
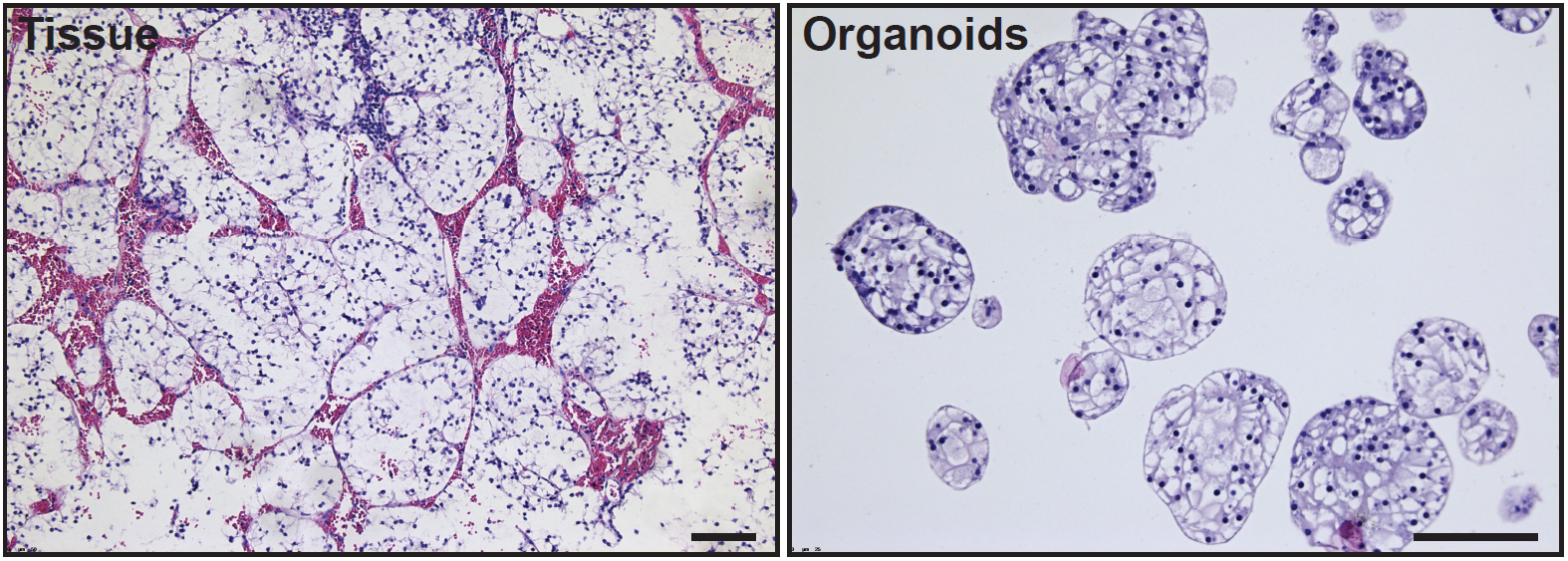
H&E stainings on clear cell renal cell carcinoma tissue (left) and organoids derived thereof (right).
Defining the developmental origin of childhood cancers
Many childhood tumors already originate in the developing fetus, but their cellular origin remains in many cases unknown. Finding the tumor’s cell-of-origin will provide important clues into what has caused its development e.g., a block in differentiation. Relieving such a block pharmacologically would be an attractive therapeutic intervention (so-called maturation therapy).
To find the origin of tumors, we make use of different approaches. First, we apply DNA sequencing technologies to find phylogenetic relations between tumors and normal tissues of the same individual. To do so, we use somatic mutations as “barcodes” to trace back the origin of the tumor. Second, we make use of (single-cell) transcriptome and epigenome sequencing approaches, as we and others have shown that embryonal tumors mirror features of their cells-of-origin to a large extend. By comparing tumor transcriptomes with the transcriptomes of fetal tissue development, we aim to unravel the tumor’s cellular identity.
Studying the (epigenetic) processes driving tumor initiation and progression
The pediatric cancer organoid models that we have established give us the unique opportunity to study the processes driving tumor initiation, progression, and therapy resistance. We are particularly interested in the epigenome changes occurring in pediatric tumors uniquely defined by mutations in SWI/SNF complex members. By using tumor organoids in combination with gene editing technologies and (single-cell) omics read-outs, we aim to pinpoint the tumor-driving signaling pathways that could serve as therapeutic targets. Furthermore, we have developed orthotopic organoid xenograft models and barcode lineage tracing strategies that allow us to study clonal dynamics during the different steps of tumorigenesis (such as primary tumor growth, metastasis formation, and therapy resistance) in great detail.
Besides tumor organoids, we use healthy tissue-derived organoids for the generation of tumor models. We apply different genome editing technologies (such as CRISPR/Cas9 technology) to healthy organoids to generate tumor progression models. Such engineered tumor organoids provide genetically defined models that allow for studying the contribution of specific genetic alterations to tumorigenesis.
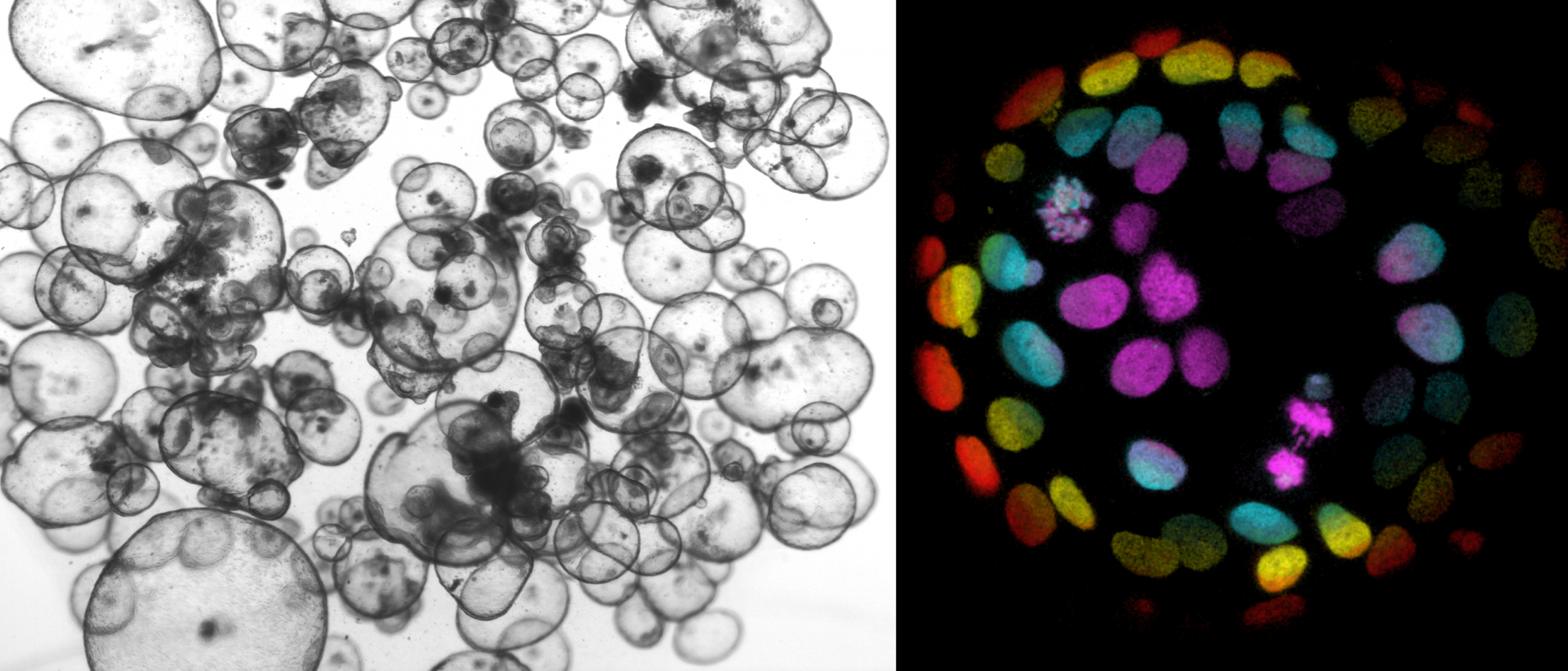
Light microscope image of healthy human colon organoids (left). Color-coded confocal image of an H2B-mNeon-expressing cancer organoid to visualize chromosome segregations (right; Drost et al., Nature 2015).
Grants
- European Research Council (ERC) Consolidator Grant (2024)
- Foundation Children Cancerfree (KiKa) Research Grant (2024, twice)
- Cancer Research Horizons CYP Therapeutic Catalyst grant (2024)
- CoFund Butterfly consortium (2023)
- Oncode Accelerator (National Growth Fund)(2023) - News Article
- Oncode Technology Development fund (2021)
- NWO-Vidi grant (2021) - News Article
- VAGABOND consortium (2021)
- Foundation Children Cancer-free (KiKa) Research Grant (2021)
- European Research Council (ERC) Starting Grant (2019) - News Article
- Foundation Children Cancer-free (KiKa) Pilot Grant (2019)
- Selected to become an Oncode Institute member (2019)
- Foundation Children Cancer-free (KiKa) Research Grant (2018)
- Foundation Children Cancer-free (KiKa) Research Grant (2017)
- NWO-Veni grant (2014)
Twinning program Máxima-KiTZ
- Validation of immunotherapy targets in rhabdomyosarcoma – with A. Banito (KiTZ)(2023)
- Drug sensitivity testing in Wilms tumor organoids – with N. Jäger (KiTZ) (2022)
- Molecular dissection of AT/RT – with M. Kool and P. Johann (KiTZ) (2021)
Princess Máxima Center Foundation
- Donations are regularly received through the Princess Máxima Center Foundation. With gratitude, the Drost group spends these donations on research with the aim to develop new, less toxic therapies for children with cancer.
Awards
- Project “The Virtual Child” shortlisted in the CRUK Grand Challenges competition (2022)
- AACR St. Baldrick’s career development award – Award for emerging leaders in the field of pediatric oncology (2020) - News Article
- Patrick Hanlo Award – Award for best post-doctoral researcher of the Hubrecht Institute (2017)
- Bas Mulder/Young Investigator Award from the Dutch Cancer Society (KWF) (2016)
Kes, M.M.G., Morales-Rodriguez, F., Zaal, E.A., de Souza, T., Proost, N., van de Ven, M., van den Heuvel-Eibrink, M.M., Jansen, J.W.A., Berkers, C.A., Drost, J.#. Metabolic profiling of patient-derived organoids reveals nucleotide synthesis as a metabolic vulnerability in malignant rhabdoid tumors. Cell Rep Med. 2025 Jan 21;6(1):101878.
Liu, N.Q.*, Paassen, I.*, Custers, L*., Zeller, P., Teunissen, H., Ayyildiz, D., He, J., Buhl, J.L., Hoving, E.W., van Oudenaarden, A., de Wit, E.#, Drost, J.#. SMARCB1 loss activates patient-specific distal oncogenic enhancers in malignant rhabdoid tumors. Nat Commun. 2023 Dec 1;14(1):7762.
DeMartino, J.*, Meister, M.T.*, Visser, L.L.*, Brok, M., Groot Koerkamp, M.J.A., Wezenaar, A.K.L., Hiemcke-Jiwa, L.S., de Souza, T., Merks, J.H.M., Rios, A.C., Holstege, F.C.P., Margaritis, T.#, Drost, J.#. Single-cell transcriptomics reveals immune suppression and cell states predictive of patient outcomes in rhabdomyosarcoma. Nat Commun. 2023 May 27;14(1):3074.
Meister, M.T., Groot Koerkamp, M.J.A., de Souza, T., Breunis, W.B., Frazer-Mendelewska, E., Brok, M., DeMartino, J., Manders, F., Calandrini, C., Kerstens, H.H.D., Janse, A., Dolman, M.E.M., Eising, S., Langenberg, K.P.S., van Tuil, M., Knops, R.R.G., van Scheltinga, S.T., Hiemcke-Jiwa, L.S., Flucke, U., Merks, J.H.M., van Noesel, M.M., Tops, B.B.J., Hehir-Kwa, J.Y., Kemmeren, P., Molenaar, J.J., van de Wetering, M., van Boxtel, R., Drost, J.#, Holstege, F.C.P#. Mesenchymal tumor organoid models recapitulate rhabdomyosarcoma subtypes. EMBO Molecular Medicine 2022 Aug 2; e16001.
Calandrini, C., van Hooff, S.R., Paassen, I., Ayyildiz, D., Derakhshan, S., Dolman, M.E.M., Langenberg, K.P.S., van de Ven, M., de Heus, C., Liv, N., Kool, M., de Krijger, R.R., Tytgat, G.A.M., van den Heuvel-Eibrink, M.M., Molenaar, J.J., Drost, J.#. Organoid-based drug screening reveals neddylation as therapeutic target for malignant rhabdoid tumors. Cell Reports 2021 Aug 24; 36(8):109568.
Young, M.D.*, Mitchell, T.J.*, Custers, L.*, Margaritis, T., Morales-Rodriguez, F., Kwakwa, K., Khabirova, E., Kildisiute, G., Oliver, T.R.W., de Krijger, R.R., van den Heuvel-Eibrink, M.M., Comitani, F., Piapi, A., Bugallo-Blanco, E., Thevanesan, C., Burke, C., Prigmore, E., Ambridge, K., Roberts, K., Vieira Braga, F.A., Coorens, T.H.H., Del Valle, I., Wilbrey-Clark, A., Mamanova, L., Stewart, G.D., Gnanapragasam, V.J., Rampling, D., Sebire, N., Coleman, N., Hook, L., Warren, A., Haniffa, M., Kool, M., Pfister, S.M., Achermann, J.C., He, X., Barker, R.A., Shlien, A., Bayraktar, O.A, Teichmann, S., Holstege, F.C., Meyer, K.B., Drost, J.#, Straathof, K.#, Behjati, S.#. Single cell derived mRNA signals across human kidney tumors. Nature Communications 2021 Jun 23; 12(1):3896.
Custers, L.*, Khabirova, E.*, Coorens, T.H.H.*, Oliver, T.R.W., Calandrini, C., Young, M.D., Vieira Braga, F.A., Ellis, P., Mamanova, L., Segers, H., Maat, A., Kool, M., Hoving, E.W., van den Heuvel-Eibrink, M.M., Nicholson, J., Straathof, K., Hook, L., de Krijger, R.R., Trayers, C., Allinson, K., Behjati, S.#, Drost, J.#. Somatic mutations and single-cell transcriptomes reveal the root of malignant rhabdoid tumours. Nature Communications 2021 Mar 3; 12(1):1407.
Calandrini, C.*, Schutgens, F.*, Oka, R., Margaritis, T., Candelli, T., Mathijsen, L., Ammerlaan, C., van Ineveld, R.L., Derakhshan, S., de Haan, S., Dolman, E., Lijnzaad, P., Custers, L., Begthel, H., Kerstens, H.H.D., Rookmaker, M., Verhaar, M., Tytgat, G.A.M., Kemmeren, P., de Krijger, R.R., Al-Saadi, R., Pritchard-Jones, K., Kool, M., Rios, A., van den Heuvel-Eibrink, M.M., Molenaar, J., van Boxtel, R., Holstege, F.C.P., Clevers, H., Drost, J.#. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nature Communications 2020;11(1):1310.
Drost, J.#, Clevers, H. Organoids in cancer research. Nature Reviews Cancer 2018 Jul, 18(7): 407-418.
Drost, J.*, van Boxtel, R.*, Blokzijl, F., Mizutani, T., Sasaki, N., Sasselli, V., de Ligt, J., Behjati, S., Grolleman, J.E., van Wezel, T., Nik-Zainal, S., Kuiper, R.P., Cuppen, E., Clevers, H. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017 Oct 13, 358 (6360): 234 – 238.
Drost, J., van Jaarsveld, R.H., Ponsioen, B., Zimberlin, C., van Boxtel, R., Buijs, A., Sachs, N., Overmeer, R.M., Offerhaus, G.J., Begthel, H. Korving, J., van de Wetering, M., Schwank, G. Logtenberg, M., Cuppen, E., Snippert, H.J., Medema, J.P., Kops, G. J. P. L., Clevers, H. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015 May 7, 521 (7550), 43 – 47.






















